How AI is Transforming Personalized Treatment: scientists are training AI to classify the effects of genetic mutations
Recently, utilizing personal information to tailor personalized treatment has gained a lot of attention. In the case of cancer, the disease begins when one or more genes in a cell are mutated. This makes each tumor distinct even if it comes from the same cell, therefore the result of a treatment may vary on different patients. With identifying genetic mutations on a patient, doctors can find out what the cause of a tumor is and give accurate treatment.
Identifying genetic mutations is becoming easier, but interpreting it remains difficult. For breast cancers, there are about 180 oncogenic mutations that contributes to the growth of tumor. To distinquish them from normal variants requires examining literature carefully. While there are thousands of publications studying the effect of genetic mutations, these information cannot be used efficiently due to lack of well-curated databases. Building such database is a costly process that requires experts to review clinical literature manually. According to Memorial Sloan Kettering Cancer Center(MSKCC), they organized an annotation committee to review data from different sources, and spent 2 months to annotate 150 genetic mutations, while there are 79 million mutations have been identified by the 1000 Genomes project. Furthermore, the number of publications is growing exponentially, so a automated classification process is in demand. To speed up the curation of mutation databases, we utilized machine’s ability to read and comprehend, which can efficiently review publications and classify the effect of genetic mutations.
From reading to comprehending
Our goal is to train a machine that can classify the mutations like human experts. Instead of training a general comprehension model, we want it to imitate the decision making procedure of experts since the amount of annotated data is insufficient.
Extract key paragraphs
Reading a 10-page paper from the beginning to the very end is time-consuming. People tend to go through the general description roughly and read the key paragraphs carefully. These paragraphs serve an important role since a sentence with mutation names in it may directly conclude the effect of the mutation. To imitate this behavior, we find keywords in the text and extract its context as key paragraphs for further investigation.
Vector representation
It is very common that words or ducuments are encoded into vector space embeddings before being processed by machine learning models. Recently, models did a great job finding representation for words. We use Word2Vec model to extract vector representation of gene names and mutation names, which are expected to be informative about its effect.
In the primary experiments, the model showed positive result as it can succesfully distinquish oncogenic mutations from normal ones. We headed to Kaggle competition [Personalized Medicine: Redefining Cancer Treatment] for a more controlled environment.
Kaggle Competition
In June 2017, MSKCC launched a Kaggle competition named “Personalized Medicine: Redefining Cancer Treatment“. The participants were asked to predict the effect of a genetic mutation given relevant documents. We collaborated with domain experts to participate this competition.
To get a basic idea of the documents, we use Doc2Vec model implemented in gensim to obtain vector space embedding of each document. The embeddings are then projected into two-dimensional space using PCA transformation. The resulting plot shows that documents from different classes can be roughly separated by its content.
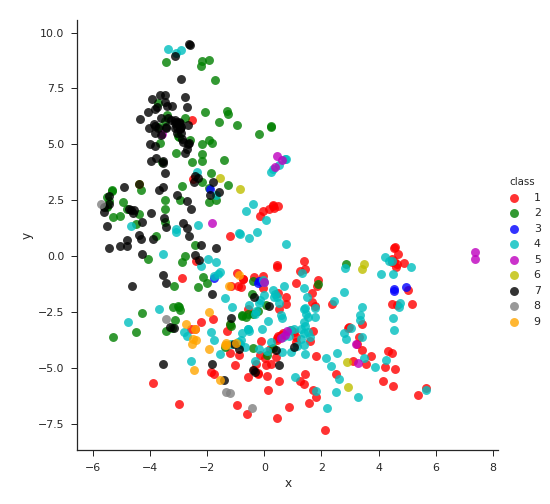
Doc2Vec embeddings with PCA transformation into 2D space
Solution to small-data problem
The dataset is relatively small with about 3000 entries; therefore we focus on feature engineering part and keep our model simple to overcome the overfitting problem. We extract classical features, such as tf-idf values, along with engineered features based on observation and domain knowledge. Keywords suggested by experts are used to extract key paragraphs, where human experts pay more attention while determining the effect of mutations.
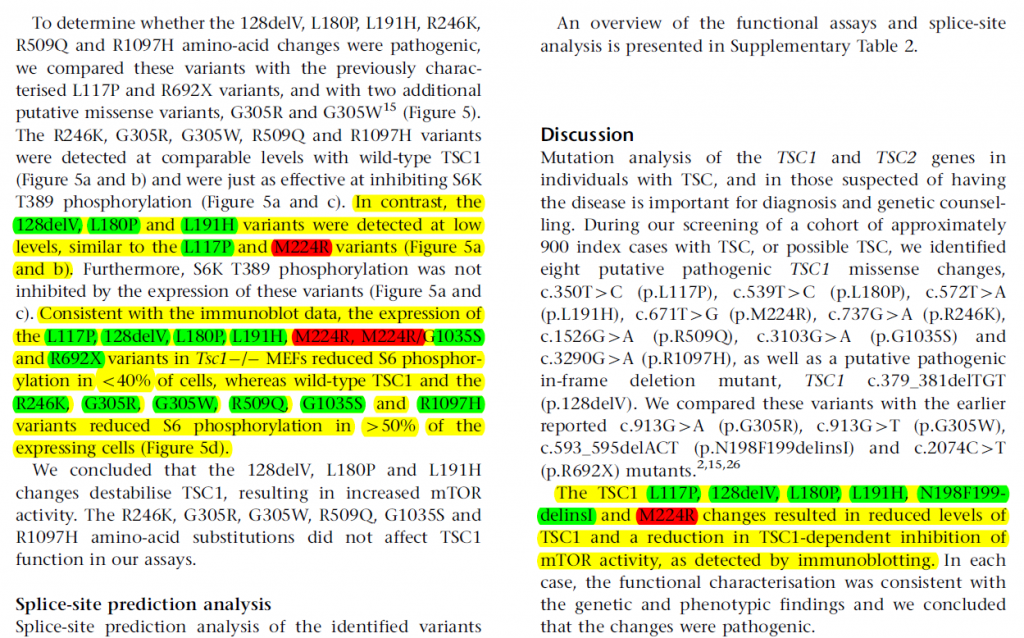
An example of key paragraph(marked yellow)
A powerful model XGBoost is used as classifier here. XGBoost has won pratically every competition in the structured data category over the last two years. In addition to strong modeling ability, its regularization technique is also well-suited for the dataset.
Result
We obtained 75% accuracy among 9 classes on the test set. The competition host also held another stage with a very different test set. In this stage, the training/testing set mismatch is too significant, makes all participants’ classifier nearly unusable.
The problem has not been well-defined. Biases in the annotating process and the ambiguity between classes has not been resolved. But the result shows that with carefully defined target, data-driven methods can be utilized to clasify the effect of genetic mutations.



Leave a Reply
Want to join the discussion?Feel free to contribute!