Extracting The Most Significant and Relevant Relational Facts From Large-Scale Biomedical Literature
Eunice You-Chi Liu
- Introduction
pubmedKB is “a novel literature search engine that combines a large number of state-of-the-art text-mining tools optimized to automatically identify the complex relationships between biomedical entities—variants, genes, diseases, and chemicals—in PubMed abstracts” (Li et al.). Currently, the pubmedKB Relation Extraction (RE) module comprises three submodules—Relational Phrases (algorithm developed by applying spaCy which is an open-source library that pioneers syntactic-dependency syntax parser), Relational Facts (model advanced by integrating R-BERT relation classification framework and BioBert) and Odds Ratio Info (tool progressed from a pre-trained general-purpose sequence-to-sequence model T5).
- Motivation
Indeed, through powerful literature mining, pubmedKB allows researchers and medical practitioners to more effectively gain extensive knowledge and insights on any biomedical entity. As this literature search engine takes all PubMed abstracts into account (currently, pubmedKB currently contains relations for 10.8M PubMed abstracts and 1.7M PubMed full texts), the relations extracted can be excessive and superfluous especially if the users want to productively identify the most pertinent relations or concisely present what relations a biomedical entity or entities pair entails. For instance, I am interested in learning the relations between HGVS:p.V600E and MESH:D009369. There are more than 2000 relations extracted for this entity pair (Figure 1). Depending on the needs of the users, scrolling over a sea of relations for this given pair might not be the most effective. Specifically, the vast majority of the relations are relational facts (denoted as rbert_cre). Moreover, when we look further into the relations extracted that are rbert_cre, there are more than 1500 relations implying similar meaning: MESH:D009369 patients that carry HGVS:p.V600E, which can be redundant and difficult to grasp what is important (Figure 2). In this case, we ask given any entity or PubMed Unique Identifier (PMID), how can we extract and demonstrate the most relevant and significant biomedical relations?
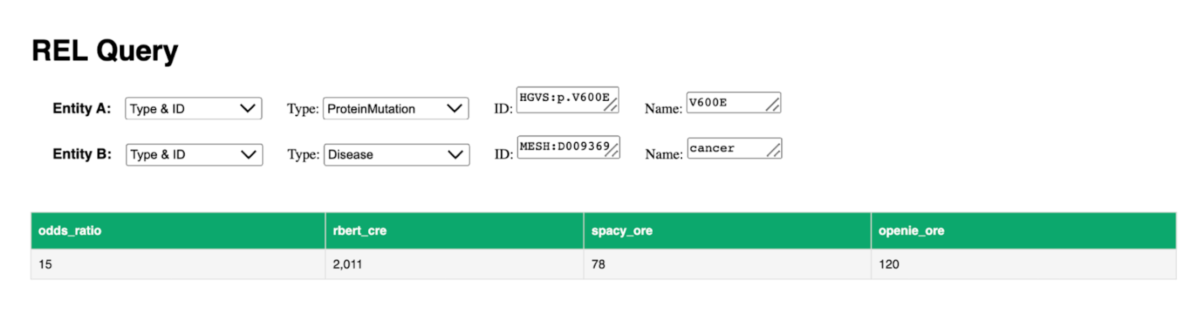
Figure 1: Using the Web GUI to search for the relations between the biomedical entities pair: HGVS: p.V600E and MESH: D009369, the number of relations found for each submodule is shown.
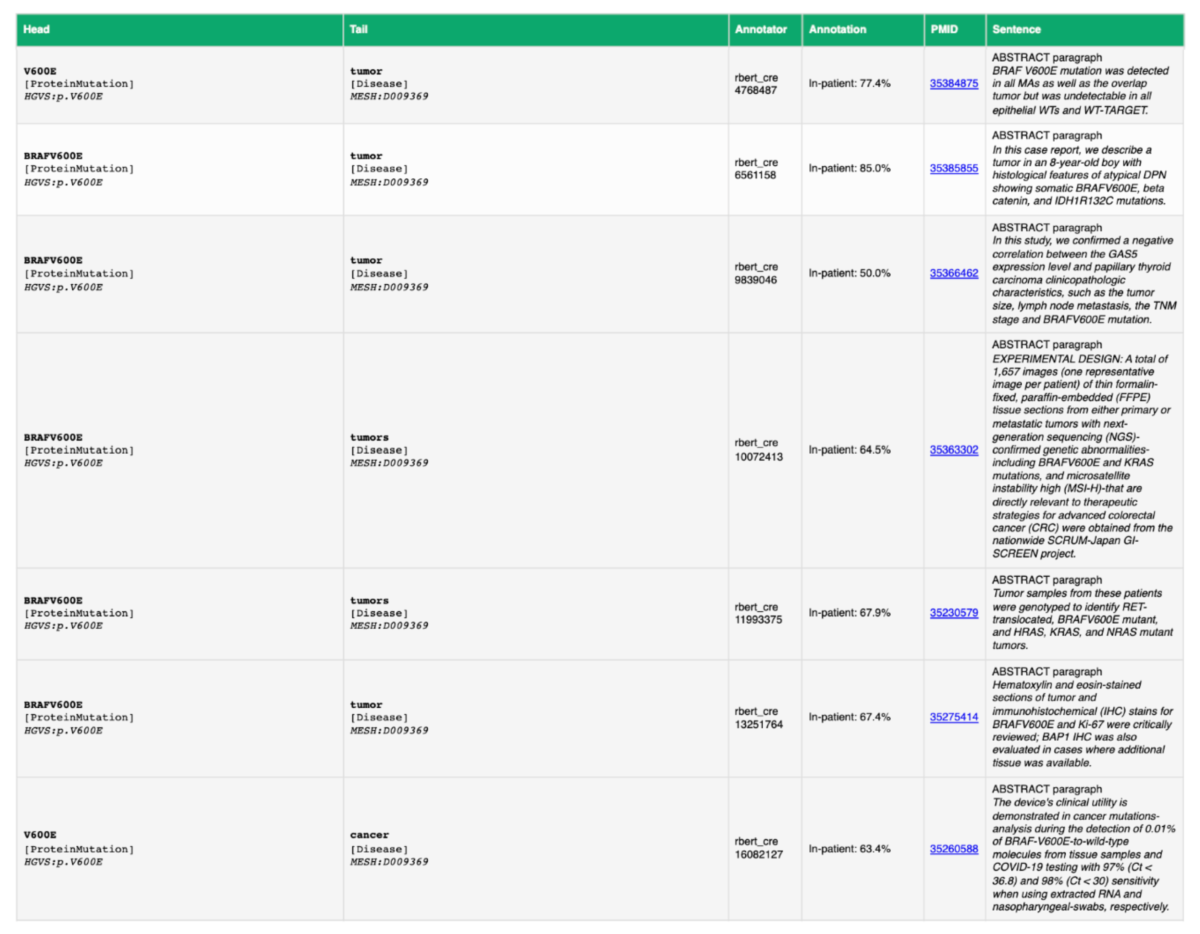
Figure 2: A demonstration of a small portion of the in-patient relations extracted between HGVS: p.V600E and MESH: D009369.
- Methods
To answer the research question posed in the motivation section, the methods can be majorly divided into two parts which are 1) “extracting” and 2) “demonstrating” the most relevant and significant biomedical relations. With the goal of acquiring the most relevant and significant relations of given entities, we develop scispaCy open relation extractions (ORE) and research on applying and developing neural open relation extractions (ORE). After finding the most accurate relations, we develop the evidence summary in order to present selective relations in a concise and informative way.
1) Develop scispaCy ORE
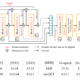
We decide to develop scispaCy ORE because it is a spaCy model specifically for biomedical text processing, which fits our main interest in working with biomedical literature and extracting significant relations from them. scispaCy dependency parsers are trained on GENIA 1.0 and then are transformed into universal dependencies, UD, format (Neumann et al.). According to spaCy’s official documentation, spaCy is trained in Clear format (English · spacy models documentation). scispaCy dependency parsers are trained on GENIA 1.0 and then are transformed into universal dependencies, UD, format (Neumann et al.). The difference in how the dependency is generated and trained might have resulted in syntactic parsing and part-of-speech (POS) tagging differences between scispaCy and spaCy (Figure 3).
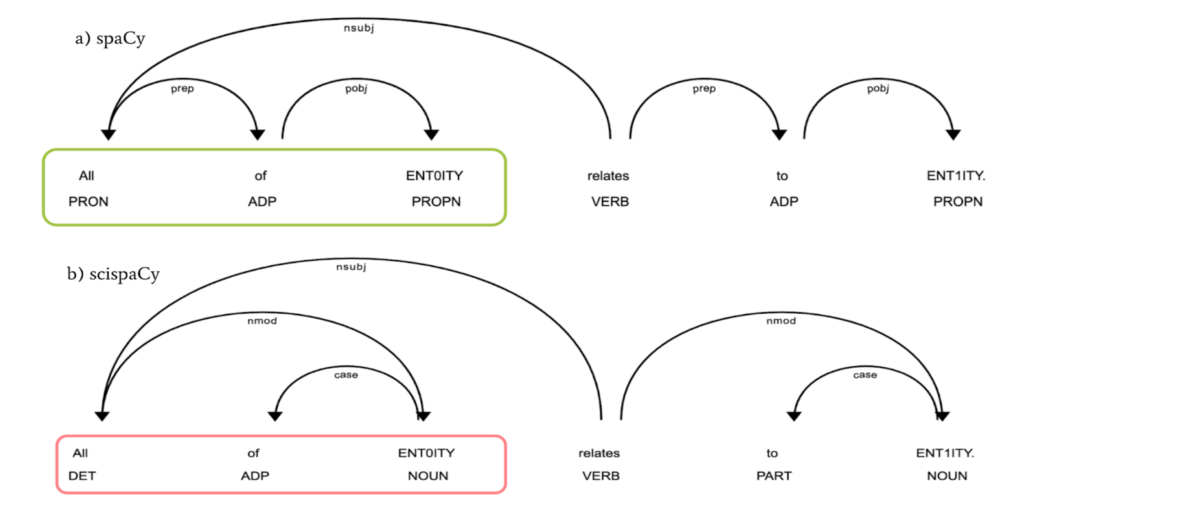
Figure 3: For a very simple sentence that entails a relation between two biomedical entities, “All of ENT0ITY relates to ENT1ITY,” in which ENT0ITY and ENT1ITY infer two different biomedical entities. The upper part (labeled as a) is the dependency parsing using spaCy and the lower part (labeled as b) is the dependency parsing applying scispaCy. Notably, the dependency parse of the same sentence (shown by arrow) and the part-of-speech (POS) tagging (marked as capitalized abbreviation under the word) are distinct in part a and b.
Using the same grammatical structure and the defined relationships between words as spaCy’s can result in misleading relations being identified. For example, as shown in Figure 3b, the main verb “relates” are directly linked to the object “ENT1ITY” rather than links to the preposition “to” in Figure 3a.
To ensure that the most relevant and important relations are extracted, understanding and redefining scispaCy’s head-dependent relationships are critical as they oftentimes “directly encode important information that is often buried in the more complex phrase-structure parses” (Jurafsky and Martin 310-334). In this case, we redefine scispaCy’s active and passive sentence structure capturing the subject, predicate, meaningful preposition, and object, and ensure that prepositions following the predicate and before entities in a noun modifier (nmod) dependency are captured. We also created a customized noun chunk extraction and set it as an extension as the default noun chunk extraction does not correspond to scispaCy’s dependency parsing. Besides the difference in dependency parsing, scispaCy and spaCy’s part-of-speech tagging diverges. In this case, we re-tag certain groups of words to ensure that the complete entities are pinpointed because for instance in Figure 3, the first entity should be “All of ENT0ITY” instead of only “ENT0ITY” because if today the determiner is changed from “all” to “none,” then the relations extracted can be inaccurate.
The relations extracted using spaCy or scispaCy ORE are called triplets as they all include one head mention, one predicate, and one tail mention. After the triplets are identified, we calculate the number of triplets extracted and randomly select 20,000 sentences with or without triplets and manually label them. In order to be counted as a valid triplet, the triplet has to match the original meaning of the sentence, should be an assertion, and be understandable (unambiguous) without looking at the original sentence.
2) Research on neural ORE
The current pubmedKB’s ORE submodule–spaCy–and the newly developed scispaCy ORE are all rule-based and can extract relations on a sentence level. As our goal is to extract the most significant and relevant relations, focusing on the sentence level might not be sufficient. We decide to research and develop neural ORE which can enable us to extract relations from paragraphs, sections, and hopefully the whole paper. Inspired by the paper “OpenIE6: Iterative Grid Labeling and Coordination Analysis for Open Information Extraction,” we adopt and applied the OpenIE6 model to our own data of PubMed abstracts and full texts. OpenIE6 model contains a series of different pipelines, which includes a BERT encoder that “computes contextualized embeddings for each word” and a 2-layer transformer that attains the contextual embeddings at every level (Kolluru et al.). Extractions, in other words, triplets, are generated at every iteration after passing in the contextualized embeddings to produce words’ labels at a specific level (Kolluru et al.).
The OpenIE6 model is trained on Wikipedia sentences, which is the same dataset as the one used for OpenIE4 (Kolluru et al.). We prepared the biomedical sentences from PubMed abstracts and full texts to get extractions. In order to evaluate the performance of the model on our biomedical sentences, we also prepared gold standard data derived from spaCy’s triplets that match the format of OpenIE6’s testing data.
3) Develop and generate the evidence summary
After gathering the most relevant and significant relations, we hope to demonstrate them in a clear and intuitive way. Figure 4 demonstrates an overview of how the evidence summary is generated. Aiming to be as diverse and informative as possible, we design that the summary generated would contain three different kinds of relations which are comprised of odds ratio, relational fact, and relational phrase via ORE. To achieve the concise aspect, we skillfully selected the relations with the highest confidence value–highest odds ratio and most supported relational phrases. In particular for the relational facts, based on the level of informativity, if the confidence value is identical, the causal relation between genetic variants and diseases is favored over the relation that entails certain patients carrying some specific diseases and the appositional relation of genetic variants and diseases. In terms of which relational phrases to feature, at most two different relational phrases from distinct sentences are included in the summary. Particularly, the relational phrases are selected based on the number of relations extracted from a single sentence as it is highly possible to infer that these sentences contain crucial information.
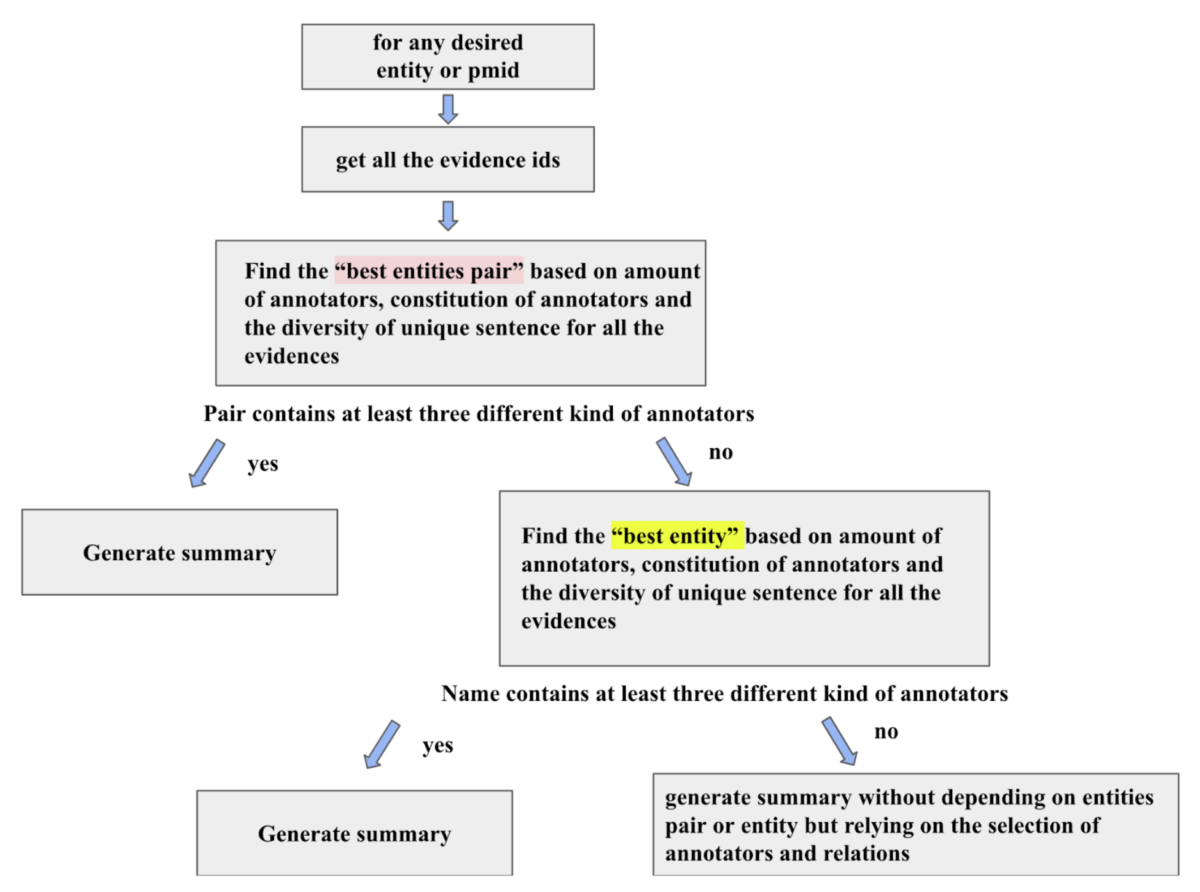
Figure 4: A quick overview and explanation of how the evidence summary is being designed and produced.
- Results
Using the newly developed scispaCy ORE, among 95,441 unique sentences extracted from various PubMed abstracts, we are able to identify and extract 1000 more triplets compared to employing spaCy ORE (Table 1).
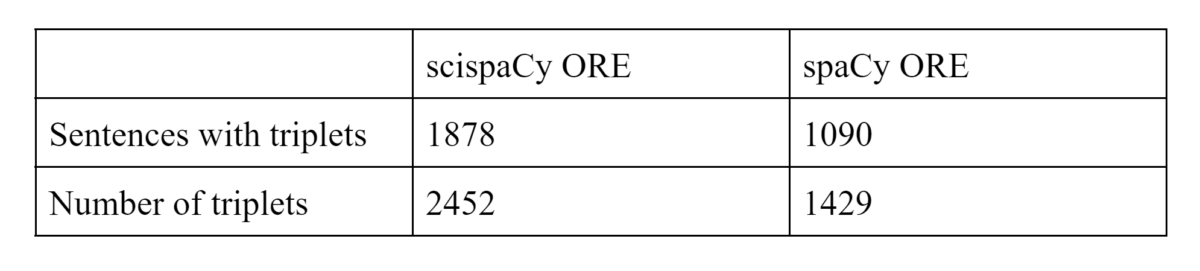
Table 1: Among 95,441 individual sentences extracted from various PubMed abstracts, the number of sentences containing identified triplets and the number of identified triplets are both higher when applying the newly developed scispaCy ORE.
We proceed into qualitative evaluation by manually looking over triplets that occurred in the randomly selected 20,000 sentences. Based on Table 2, by all means, the yield when applying scispaCy ORE is higher because a larger number of triplets are identified. The precision of the correct triplets (those who fit the criterion mentioned in the method section) extracted from scispaCy and spaCy is similar (Table 2). We can not really calculate recall because it would be nearly impossible to identify false negative triplets.
Furthermore, we discover that scispaCy ORE allows us to generate triplets in sentences that have possessive relationships and adjective comparisons which current spaCy ORE is not yet able to achieve. To illustrate, for the sentence “Our results indicate that CYFRA 21-1 may be a useful tumor marker in NSCLC, especially in carcinoma planocellulare,” the triplet–“CYFRA 21-1, marker in, NSCLC”–is identified and extracted using scispaCy ORE but not spaCy ORE. In this case, scispaCy ORE is capable of identifying and incorporating relations other than action verbs and these other relations are also common and critical in biomedical literature besides those focused on an action verb.
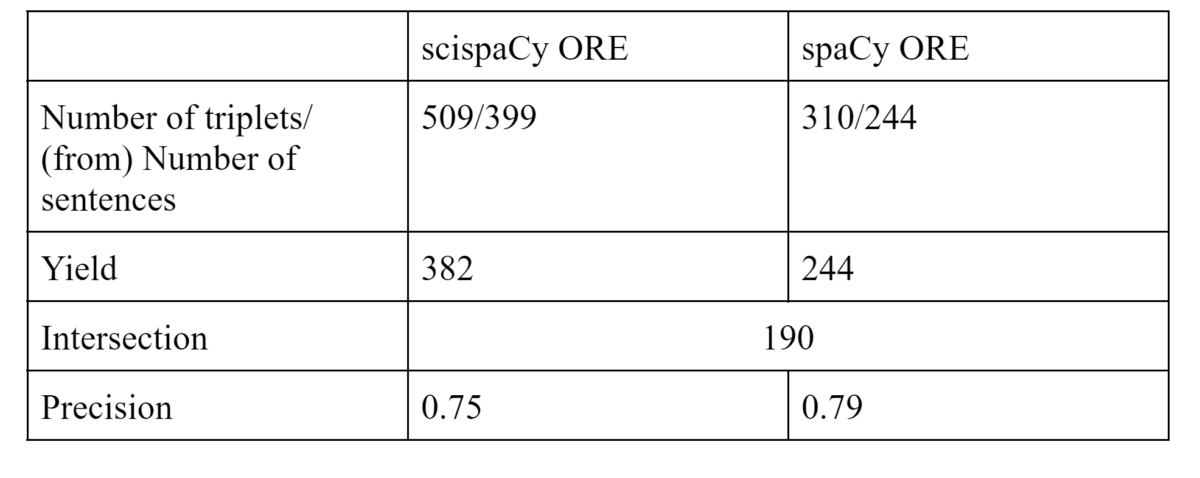
Table 2: After randomly selecting 20,000 sentences and labeling them, the number of correct triplets extracted in scispaCy is 382 in spaCy is 244. Among the number of correct triplets extracted in scispaCy ORE and spaCy ORE, 190 triplets are identical. The precision of the correct triplets extracted from scispaCy and spaCy is similar, 0.75 and 0.79 respectively.
Besides having scispaCy ORE that extracts and identifies important triplets which are missing in spaCy ORE, we recognize that the performance of neural ORE, specifically the OpenIE6 model, is also promising. By creating one-to-one mapping between the predicted extractions (generated by the OpenIE6 model) and the spaCy original extracted triplets, we evalutate the precision and recall. Notably, the AUC and F1 score, 49.0 and 62.9, are comparatively the same as the testing results using the standard CaRB dataset –a large-scale Open IE benchmark annotation that aims to contribute to the standardization of Open IE evaluation (Stanovsky and Dagan).
After developing and researching the most suitable ORE to extract the most significant and relevant relations, we compiled those information into a well-organized and informative paragraph (Figure 5). Moreover, the relations annotated by each annotator, including odds ratio, relational phrase, and relational fact are clearly labeled in distinct colors for easier comprehension (Figure 5).

Figure 5: The summary of the relations between HGVS: p.V600E and MESH: D009369.
- Discussion
In this study, we ask and answer the question: given any entity or PubMed Unique Identifier (PMID), how can we extract and demonstrate the most relevant and significant biomedical relations? We add on to previous work especially to the development of pubmedKB by developing a new biomedical-specific sicspaCy ORE and researching the application of neural ORE. Moreover, we organize and present those selected relations into a well-informative paragraph ready for the usage of demonstrating in academic and professional contexts. Future studies can further expand on the development of neural ORE given the positive performance of the OpenIE6 model. Currently, the OpenIE6 model is trained on sentences from Wikipedia. In this case, training the OpenIE6 model with both biomedical sentences from PubMed and general sentences from Wikipedia can potentially yield greater performance. In terms of the scispaCy ORE, although it is a comparatively developed and mature submodule ready to be used. It might also be interesting to explore scispaCy’s named-entity recognition (NER) module and see how it can be integrated into the ORE.
Works Cited
“Dependency Parsing.” Speech and Language Processing: An Introduction to
Natural Language Processing, Computational Linguistics, and Speech
Recognition, by Dan Jurafsky and James H. Martin, Pearson, 2022, pp.
310–334.
“English · Spacy Models Documentation.” English,
spacy.io/models/en?fbclid=IwAR2_DCNpcRx8qp_iB0b5_QdTjljmL2oty
pUMTLm4p-wGP_8CYuvLnlfH6os#en_core_web_sm.
Kolluru, Keshav, et al. “OpenIE6: Iterative Grid Labeling and Coordination
Analysis for Open Information Extraction.” Proceedings of the 2020
Conference on Empirical Methods in Natural Language Processing
(EMNLP), 2020, doi:10.18653/v1/2020.emnlp-main.306.
Li, Peng-Hsuan, et al. “PubmedKB: An Interactive Web Server for Exploring
Biomedical Entity Relations in the Biomedical Literature.” Nucleic Acids
Research, vol. 50, no. W1, 2022, doi:10.1093/nar/gkac310.
Neumann, Mark, et al. “ScispaCy: Fast and Robust Models for Biomedical
Natural Language Processing.” Proceedings of the 18th BioNLP
Workshop and Shared Task, 2019, doi:10.18653/v1/w19-5034.
Stanovsky, Gabriel, and Ido Dagan. “Creating a Large Benchmark for Open
Information Extraction.” Proceedings of the 2016 Conference on
Empirical Methods in Natural Language Processing, 2016,
doi:10.18653/v1/d16-1252.